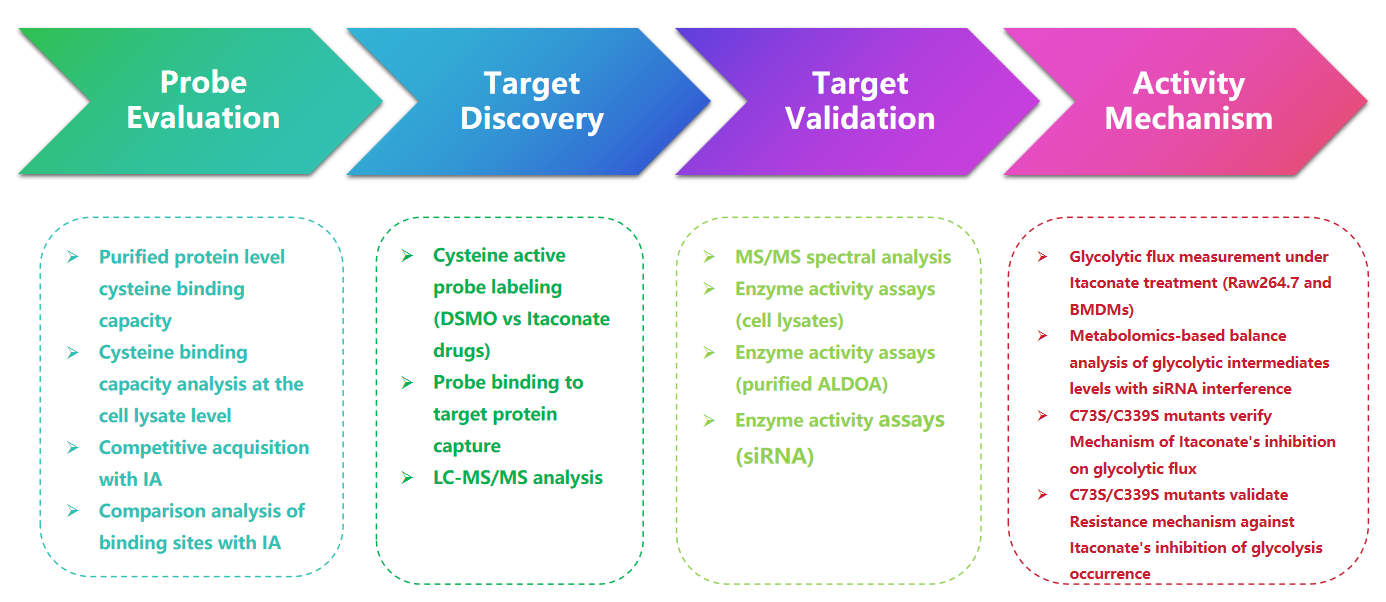
ABPP analysis reveals new anti-inflammatory mechanism of itaconic acid via protein-metabolite interactions
Itaconate, an endogenous metabolite crucial for inflammation and immune regulation, has its role elucidated in a joint paper by the ChomiX Biotech team. The study innovatively used ABPP technology to reveal that itaconate modifies S-glycosylation of cysteine residues on key glycolytic enzymes, thereby impacting cellular metabolism. Researchers mapped protein-itaconate interactions, finding that itaconate directly binds and regulates multiple enzymes in the glycolysis pathway, affecting its rate and direction. This research advances our knowledge of metabolic control mechanisms during disease-related inflammation and demonstrates ABPP's strength in exploring how small molecules modulate protein functions. As a leader in chemical proteomics, Corolus BioScience offers comprehensive solutions, including probe design, sample processing, high-throughput screening, and data analysis, to help clients uncover similar metabolic-regulatory mechanisms.
1. Detection of Itaconate Modifications with the 1-OH-Az Probe
In this study, the authors employed advanced profiling techniques to investigate itaconate-targeted cysteine residues. They initially assessed probe 1-OH-Az through gel electrophoresis and performed competitive experiments using IA-alkyne. Mass spectrometry validation confirmed that 1-OH-Az selectively labels cysteine sites, with 87% of newly identified proteins not previously documented as ligand-binding proteins in DrugBank. Given the diverse functions and disease relevance of these proteins, the 1-OH-Az probe can be used to identify active cysteines as potential drug targets for novel therapeutics.
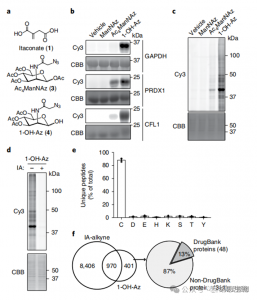
Figure 1: 1-OH-Az as an Efficient and Distinctive Cysteine Analysis Probe for Detecting Itaconate Modifications
2. Identification of Itaconate Modification Sites Using Quantitative Proteomics Techniques
The researchers conducted isoTOP-ABPP experiments to quantify cysteine residues modified by itaconate using the 1-OH-Az probe. After lysate pretreatment, isotopically labeled linker-assisted quantitative proteomics were utilized to analyze the impact of varying concentrations of itaconate, pinpointing its specific targets. Comparative experiments were also done using two concentrations of IA-alkyne, which, while showing broader activity and coverage, only recognized 65 and 50 effective competing sites. Notably, 1-OH-Az demonstrated a significantly higher competitive advantage among co-quantified cysteines.
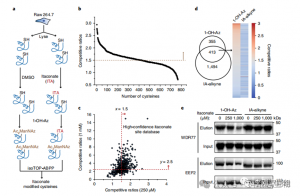
Figure 2: Chemical Proteomic Analysis of Itaconate-Modified Cysteines Utilizing Competitive isoTOP-ABPP with 1-OH-Az
3. Itaconate Modifies and Inhibits a Key Glycolytic Enzyme
Mass spectrometric analysis revealed that itaconate modifies three key glycolytic enzymes: ALDOA, GAPDH, and LDHA. Endogenous itaconate modification on Cys73 and Cys339 of ALDOA was confirmed within LPS-stimulated Raw264.7 cells. Due to their proximity, the authors speculated that such modifications might influence aldolase activity. Indeed, treatment with 1 mM itaconate led to reduced ALDOA enzyme activity without affecting protein expression. Furthermore, isoTOP-ABPP analyses showed that Cys84 on LDHA and Cys245 on GAPDH are also targets for itaconate modification.

Figure 3: Itaconate Can Modify and Impair ALDOA Function
4. Itaconate Mainly Suppresses Glycolysis by Targeting ALDOA
To assess ITAC's regulatory role in inflammatory macrophage glycolysis, the authors monitored glucose consumption and lactate production in Raw264.7 cells before and after LPS stimulation, demonstrating that ITAC significantly decreased both, indicating suppression of glycolytic function. To validate ITAC's effect on ALDOA and subsequent inhibition of glycolysis, they knocked down endogenous ALDOA using RNAi and overexpressed either WT or double-mutant (C73S/C339S) ALDOA in Raw264.7 cells. As expected, knockdown of ALDOA resulted in decreased glucose consumption and lactate production, rendering the cells insensitive to ITAC treatment. Reintroduction of WT or mutant ALDOA restored metabolic levels in untreated cells; however, cells overexpressing the mutant ALDOA exhibited reduced sensitivity to glycolytic inhibition compared to WT. Enzymatic assays for aldolase activity aligned with glycolytic states.
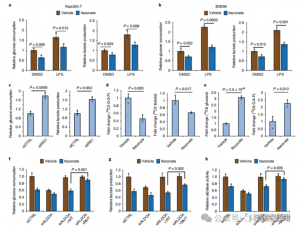
Figure 4: Itaconate Alters the Glycolytic Pathway via Modifying ALDOA
5. Inhibition of ALDOA Contributes to Anti-Inflammatory Responses
These findings unveiled that ITAC suppresses glycolytic pathway activity by modifying cysteine residues Cys73 and Cys339 on ALDOA. The authors further inferred from the known anti-inflammatory action of dimethyl fumarate via cysteine modification on GAPDH and glycolytic inhibition that ITAC may similarly feedback to interfere with glycolysis to exert an anti-inflammatory effect. Knockdown of ALDOA significantly reduced IL-1β secretion upon LPS stimulation, suggesting its involvement in regulating inflammation through glycolysis. Partially reversing the anti-inflammatory effects of ALDOA knockdown with supplemented pyruvate indicated a limited role for LDHA in inflammatory responses.
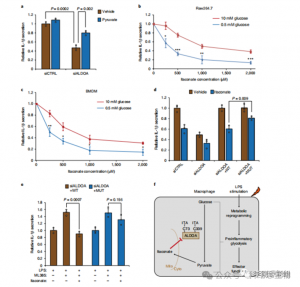
Figure 5: The Anti-Inflammatory Action of Itaconate is Mediated through Its Inhibition of ALDOA, Resulting in Disruption of Glycolysis
In summary, this paper effectively harnessed S-glycosylation-based cysteine profiling technologies and ABPP methodologies to construct a network of interactions between itaconate and related proteins. It convincingly demonstrates that itaconate serves as a critical metabolic regulator by specifically modifying cysteine residues on proteins to control the glycolytic pathway. This research not only elucidates the mechanism of action of itaconate as a novel metabolic regulator but also provides robust evidence for how small metabolites interact with proteins to modulate core metabolic pathways.
In addition to ABPP, other methods for studying the interactions between small-molecule metabolites and proteins include, but are not limited to:
1. Affinity enrichment (pull-down) techniques, where the native compound is converted into a biotinylated probe that incubates with cell lysates, followed by streptavidin-mediated enrichment and isolation of protein targets that bind to the biotin probe.
2. Limited proteolysis mass spectrometry (LiP-MS) is a target discovery technology based on protein affinity. When specific ligands, such as drugs or small molecules, bind to particular proteins, they induce conformational changes or steric hindrances leading to differential cleavage sites compared to the unbound protein. By detecting these differences using mass spectrometry, this method can identify drug-protein interactions within cells and determine the drug's molecular targets.
3. Cellular Thermal Shift Assay (CETSA) was initially developed to aid in anticancer drug target research and is one of the first widely used label-free methods for studying drug-target engagement in intact cells. CETSA primarily relies on the principle that upon binding to a target protein, a compound increases its thermal stability. After incubating samples with the compound and corresponding controls at various temperature gradients, proteins bound to the ligand remain folded and relatively stable following heating, whereas unbound proteins unfold and precipitate due to denaturation. Subsequent analysis of soluble protein thermostability through immunoblotting or mass spectrometry-based approaches, based on their melting curves, confirms the interaction between the compound and intracellular proteins.
4. Affinity chromatography involves conjugating the target metabolite to a solid-phase matrix to capture protein complexes that interact with it. The captured proteins are then identified using techniques such as mass spectrometry.
5. Surface Plasmon Resonance (SPR) is used for real-time, label-free measurement of kinetic parameters associated with the interactions between small molecules and proteins.
6. Protein crystallography elucidates the three-dimensional structures of proteins complexed with small molecule ligands, providing an intuitive insight into the binding sites and mechanisms of action. By solving these structures, researchers can directly visualize how the small molecules interact with their protein targets.