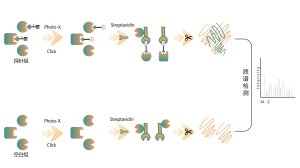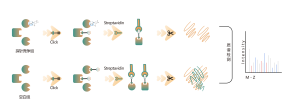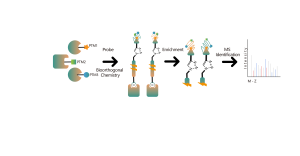
Products
Small Molecule-Protein Interactions
Proteins, as the direct participants and executors of life activities, represent crucial targets for disease therapy. Small molecule drugs (organic compounds typically with a molecular weight less than 1000 Da) exert effective therapeutic effects by finely modulating protein activities, abundances, and interactions. Common small molecule drugs include natural products and their derivatives (e.g., herbal monomers) as well as chemically synthesized drugs. Upon entering the human body, these drugs exert their therapeutic effects by binding to target proteins within cells or externally. Therefore, understanding how small molecule drugs bind to target proteins is particularly crucial in drug development, especially in complex physiological environments such as living cells, blood, and diseased tissues. In-depth analysis of the interaction between small molecule drugs and proteins not only enables precise identification of drug targets but also reveals the molecular mechanisms of drug action and potential off-target effects. Moreover, it holds the promise of discovering novel therapeutic targets, thereby providing richer strategies for disease treatment.
In addition to small molecule drugs, endogenous small molecule metabolites within organisms, such as ATP, cholesterol, bile acids, arachidonic acid, and retinoic acid, participate in regulating many important signaling pathways and protein activities by interacting with proteins, including transport proteins, membrane receptors, transcription factors, and metabolic enzymes. In recent years, the interaction between gut microbiota metabolites and host cells has emerged as a research hotspot. Therefore, thorough investigation and mapping of the interaction networks between metabolites and proteins in cellular environments, especially under diseased states, are of significant importance for understanding life processes and treating diseases.
Chemical proteomics, as a significant branch of chemical biology, has now been widely employed in protein function research, small molecule drug target identification, and new drug structure screening. This technological platform utilizes a variety of functionally diverse chemical probes, combined with proteomics, aiming to elucidate the interaction mechanisms between small molecules and proteins under physiological conditions (such as living cells, blood, tissues, etc.). It's worth mentioning that, compared to purified protein systems, the use of living cell systems is a major characteristic of chemical proteomics. It allows for a realistic depiction of the distribution of targets for small molecule drugs, endogenous metabolites, etc., within complex proteomes, even down to the level of amino acid residue sites.
Chomix offers professional analysis services for small molecule-protein interactions, empowering you to delve deeper into potential drug targets and enhance your understanding of drug molecular mechanisms and potential side effects. Our expert team possesses extensive experience in chemical proteomics research and will select the most suitable and reliable methods for you, eliminating any concerns about technical challenges and facilitating the advancement of your research effortlessly.

Technical Service
1. Identification of Direct Targets for Non-covalent Small Molecule Drugs
Most small molecule drugs interact with target proteins through non-covalent binding, forming dynamic and reversible interactions with amino acid residues in binding pockets via hydrogen bonds, π-π stacking, hydrophobic interactions, etc. Therefore, stable enrichment and isolation of proteins bound by non-covalent small molecule drugs from complex proteomes pose significant challenges. To address this, Chomix has developed a chemical proteomics target identification platform based on photoprobes. This platform accurately captures the dynamic binding between small molecules and proteins in living cells and achieves separation and enrichment, thereby comprehensively identifying direct targets for non-covalent small molecule drugs at the proteomic level.
2. Identification of Binding Pockets for Non-covalent Small Molecule Drugs
In the process of small molecule drug development, it is crucial to first determine the binding information between the drug and the protein, and then specify which particular pocket on the protein surface the drug binds to and how it binds. This information is essential for subsequent drug structure optimization. In addition to classical structural biology approaches, Chomix has also developed an advanced chemical proteomics technology platform based on high-resolution mass spectrometry. This platform can identify binding peptides for non-covalent small molecule drugs at the protein and even cellular levels, thereby assisting in addressing this critical issue in early drug development.
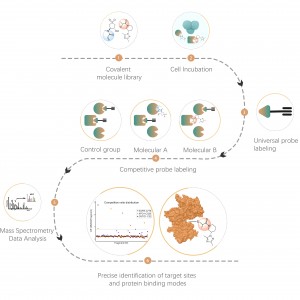
3. Quantitative Analysis of Occupancy and Selectivity of Covalent Small Molecule Drug Targets
Covalent drugs refer to drugs that form stable covalent bonds with amino acid residues at binding pockets on target proteins, such as those binding to cysteine, lysine, serine, etc. Common covalent drugs include aspirin, osimertinib, zebularine, as well as natural products like artemisinin and artesunate. In living cells, covalent drugs can stably bind to and occupy specific amino acid residues on target proteins. Leveraging this characteristic, Chomix has successfully developed a chemical proteomics platform based on a universal probe. This platform enables quantitative analysis of target site occupancy for covalent small molecule drugs, down to the level of amino acid residues. Moreover, by analyzing occupancy information for over 10,000 amino acid residue sites, it can determine target selectivity at different drug concentrations, providing powerful guidance for early drug development.
4. Identification and Selectivity Analysis of Protein Degrader Targets
As a novel type of drug, Proteolysis Targeting Chimeras (PROTACs) differ from traditional small molecule inhibitors or activators. They subvert the conventional "occupancy-driven" development concept in medicinal chemistry by utilizing the endogenous ubiquitin-proteasome system (UPS) to specifically degrade disease-causing proteins, especially those considered "undruggable" targets. Therefore, quantitative identification of protein degrader drug targets and their selectivity at the whole proteome level is crucial for early development of such drugs. Chomix has successfully developed various quantitative chemical proteomics technology platforms capable of qualitatively and quantitatively analyzing over 5,000 proteins in individual cell lines, providing comprehensive and in-depth analysis of target selectivity.